- His12 acts as a general base, accepting the 2'-OH proton of the sessile ribonucleic sugar ring. This promotes a nucleophilic attack by the 2'-O on the more positively-charged phosphorus (P) atom thereby creating a 2’,3’-cyclic ribonucleotide phosphate intermediate. The creation of this intermediate is facilitated by the imidazole of His119 which acts as a general acid, donating its proton to the oxygen atom in the susceptible P-O-R' bond.
- For the second step of the reaction, the general acid and base activities of the sidechains of these two His residue are reversed. His12 acts as a general acid, donating its newly acquired proton to the 2’,3’-cyclic ribonucleotide phosphate intermediate, while His119 which acts as a general base, accepting a proton from water to promote hydroxyl attack on the same 2’,3’-cyclic intermediate.
Uridine vanadate is a potent competitive
inhibitor of RNase A. What makes it so potent is the fact that its structure mimics a transition state intermediate in the reaction. Specifically, the oxidized vanadium atom is
pentavalent ion with five coordinated oxygen atoms whose stable structure is believed to
mimic the bipyramid structure of an unstable pentavalent phosphodiester intermediate that
transiently forms in the active site of the
enzyme.
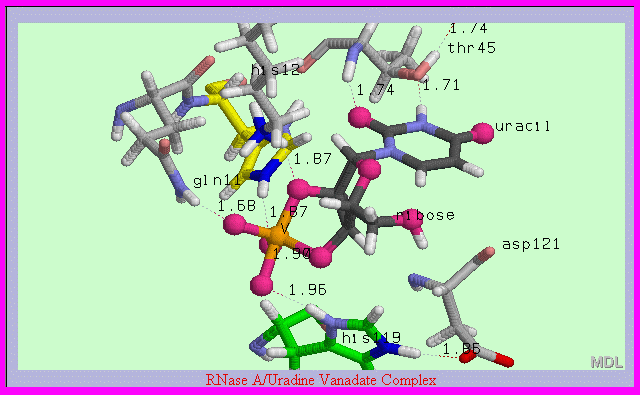 |
Legend: 2',3'-cyclic uridine
vanadate complex with RNase A active site: The pentacovalently coordinated vanadium (V) atom of the inhibitor forms metal bonds with 5 oxygen
atoms (red spheres)
creating a bipyramidal 2',3' cyclic ribonucleotide (black wireframe) linked to uracil.
Specific active site residue contacts with His 119, His12, Lys41, and Thr45 are shown with
distance measurements indicated in Ångstroms.
| Go to top | |
| © | Duane W. Sears Revised: October 07, 2010 |